ASP (Domoic acid)
direct ELISA
Enzyme immunoassay for the quantitative determination of amnesic
shellfish poison (Domoic acid) in algal, shellfish or seawater extracts.
 BS31300401
BS31300401
 96
96
For illustrative purposes only.
To perform the assay the instructions for use provided with the kit have to be used.
Distributed by:
I B L I N T E R N A T I O N A L G M B H
Flughafenstrasse 52a Phone: +49 (0)40-53 28 91-0 IBL@IBL-International.com
A S P E L I S A K I T
FOR QUANTITATIVE DETERMINATION OF
DOMOIC ACID IN SHELLFISH
PROD. NO.: A31300401 P R E - R E L E A S E
A . I N T R O D U C T I O N
Domoic acid (DA) and DA derivatives are water-soluble neurotoxins produced by
a number of marine algae, in particular by the microalgae of the genus Pseudonitzschia
spp (Fig. 1). Blooms of Pseudo-nitzschia may lead to the accumulation
of DA in shellfish filter feeders and other marine species [Scholin et al., 2000].
Ingestion of DA contaminated shellfish may lead to amnesic shellfish poisoning
(ASP) by affecting the central nervous system, and has caused the death of both
animal and human consumers in severe cases [Wright et al., 1989]. The European
commision Directive 2002/226/EC implemented a maximum permitted level (MPL)
of 20 mg DA equivalents/kg shellfish intended for human consumption. This MPL
is adopted by most food safety authorities.

F I G U R E 1 : D O M O I C A C I D S T R U C T U R E
Enzyme Linked Immunosorbent Assay (ELISA) has proved to be a sensitive and
rapid method for detection of DA in the marine environment [Garthwaite et al.,
2001]. This quantitative DA ELISA was developed by AgResearch (Hamilton, New
Zealand) for the detection of DA in water samples, shellfish and algal extracts, and
is based on antibodies described by Garthwaite et al., 1998. The assay is specific
for DA, with no cross-reactivity to non-toxic, structural analogues like kainic acid,
L-glutamic acid, L-glutamine, formimino-L-glutamic acid, proline or g-aminobutyric
acid (GABA). The assay is primarily intended for use in routine monitoring of DA
levels in bivalve molluscs to comply with the regulatory MPL, but is also applicable
for DA quantification in other marine matrixes. The ASP ELISA has been subjected
to comprehensive single-laboratory and inter-laboratory validation studies
(Biosense report 2003:6 and 2004:1.2).
Assay principle
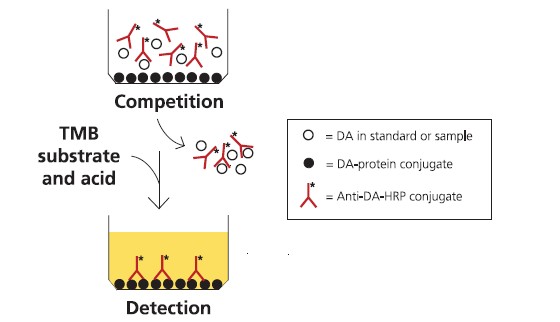
The ASP ELISA assay is in a direct competition format, where free DA in the
sample competes with DA-conjugated protein coated on plastic wells for binding
to anti-DA antibodies free in the solution (Fig. 2). The polyclonal ovine anti-DA
antibodies are conjugated to horseradish peroxidase (HRP). Sample diluted in
buffer is incubated in the wells with the anti-DA-antibody-HRP conjugate. After
washing, the amount of conjugate remaining bound to the well is measured by
incubation with a substrate that gives a blue product upon reaction with the HRP
enzyme. Addition of acid stops the reaction and changes the product colour from
blue to yellow. The colour intensity is measured spectrophotometrically on a platereader
at 450 nm, and is inversely proportional to the concentration of DA in the
sample solution. The assay is calibrated using dilutions of a DA calibration solution
supplied with the kit. The calibrated range of the assay (I(20) – I(80)) is approximay 10
to 300 pg/mL of DA. The ASP ELISA kit can either be used in 2 rounds of analysis for
the quantification of 12 individual sample dilutions in duplicate (plus calibration
solutions, A(max) and blanks), or for the quantification of 36 sample dilutions in
duplicate using all 8 strips. The working range for ASP toxins in shellfish is 0.01mg/
kg up to at least 200 mg/kg.
F I G U R E 2 : A S S A Y F O R M A T F O R T H E C O M P E T I T I V E A S P E L I S A
B . S A F E T Y I N S T R U C T I O N S
As all chemicals should be considered potentially hazardous, always wear suitable
protective clothing during handling of the kit.
CAUTION: Domoic acid is a neurotoxin that is harmful by inhalation and ingestion.
Avoid contact with skin, eyes and clothing. Wash hands thoroughly after
handling.
Beware of the hazardous nature of methanol and sulfuric acid. Please refer to the
manufacturers Material Safety Data Sheet for these reagents.
C . S T O R A G E A N D S T A B I L I T Y
Store the kit at 2-8°C upon arrival. Do not freeze. See expiry date on the kit box
for stability of the kit.
D . W A R R A N T Y A N D L I M I T A T I O N O F R E M E D Y
Biosense Laboratories AS (hereafter: Biosense) makes no warranty of any kind, expressed or
implied, including, but not limited to, the warranties of fitness for a particular purpose and
merchantability, which extends beyond the description of the chemicals on the face hereof,
except that the material will meet our specifications at the time of delivery.
Buyer’s exclusive remedy and Biosense´s sole liability hereunder shall be limited to refund of
the purchase price of, or at Biosense´s option, the replacement of, all material that does not
meet our specifications. Biosense shall not be liable otherwise or for incidental or consequential
damages, including, but not limited to, the costs of handling.
Said refund or replacement is conditioned on Buyer giving written notice to Biosense within
thirty (30) days after arrival of the material at its destination, and Buyer treating the material
as outlined in the product data sheet and/or kit insert after arrival. Failure of Buyer to give said
notice within said thirty (30) days, or failure of Buyer in treating the material as outlined in the
product data sheet and/or kit insert shall constitute a waiver by Buyer of all claims hereunder
with respect to said material.
The responsibility of all patent considerations in the use of our products rests solely with the
user.
E . K I T C O N T E N T S
Number:
A) 12-well microplate strip modules 2 sealed pouches - 4 strips each
(Precoated with DA-protein conjugate)
B) Plate sealers 2
C) PBS/Tween tablets 2
D) Domoic Acid standard, 100 ng/mL 2 vials
(derived from NRC CRM-DA-e)
E) Anti-DA-HRP conjugate, 2 vials
(6x concentrated)
F) Ovalbumin 2 vials
G) TMB peroxidase substrate 2 vials
F . ADDITIONAL REAGENTS AND EQUIPMENT REQUIRED
In addition to the reagents supplied with the kit, the following reagents and
equipment are required and/or recommended to perform the assay:
• Kitchen blender.
• Centrifuge.
• Vortex mixer.
• Micropipettes.
• Microplate spectrophotometer equipped with a 450 nm filter.
• Water; distilled and deionised (e.g. Milli-Q water, Millipore).
• Methanol (analytical grade).
• 0.3 M H2SO4.
G . IMPORTANT NOTES
1. Read the complete procedure before starting the assay.
2. Protect vials and microwell strips containing DA standard dilutions and samples
from direct light during incubations.
3. The plate sealers are used to seal the strips during incubation and care must
be taken when removing them from the strips.
4. Positive displacement pipettes (50 μL) are recommended for dispensing
methanolic extracts.
5. As in every quantitative ELISA, consistent and precise pipetting at each step in the
procedure is essential in order to obtain reliable results.
6. Reproducibility in any ELISA is also dependent upon consistent washing of the
microwells.
7. After each wash, the wells are emptied by inverting the strips over a sink and
then tap dry the wells against a pile of paper towels to remove all of the remaining
liquid.
8. Avoid prolonged intervals between the working steps of the procedure, and do
not allow the microwells to dry out totally during the assay procedure.
Definitions
Blank wells: Background absorbance of the TMB peroxidase substrate;
approximay 0.05 A.U. (Absorbance Units).
Amax wells: Maximum absorbance; no standard or sample is added to these wells
allowing maximum binding of the anti-DA-HRP conjugate to the plate-coated DAconjugate;
approximay 1.0 A.U. (Absorbance Units).
H . PREPARATIONS BEFORE THE ANALYSIS
a) Preparation of buffers and reagents
1. Washing buffer (PBS-T; 0.05% Tween 20 in PBS):
Dissolve one tablet (C) in distilled water and dilute to 500 mL. May be stored
at 4ºC for one week.
2. Extraction solution (50% methanol in water):
Prepare sufficient solution for the required number of samples by mixing equal
volumes of methanol and distilled water. Prepare fresh each day.
3. Standard/Sample buffer (10% methanol in PBS-T):
Mix 5 mL of methanol with 45 mL of Washing buffer. May be stored for 2-3
days at room temperature.
4. Antibody-HRP buffer (1% ovalbumin in PBS-T):
Add 6 mL of Washing buffer to 60 mg of ovalbumin (vial F). Prepare fresh for
each assay.
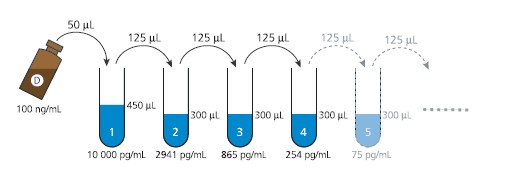
b) Preparation of Domoic acid calibration solutions
The 10-point calibration curve is freshly prepared using standard dilutions in the
range of 10 000 – 0.16 pg DA/mL:
1. Prepare one Eppendorf tube containing 450 μL Standard/Sample buffer (10%
methanol in PBS-T) - “tube 1”, and 9 Eppendorf tubes containing 300 μL
Standard/Sample buffer - “tubes 2-10”.
2. Add 50 μL of the DA standard (100 ng/mL, vial D) to tube 1 and vortex, to
obtain a 10 ng/mL DA solution.
3. Transfer 125 μL of the 10 ng/mL solution (tube 1) to tube 2 and vortex.
4. Complete the 3.4-fold dilution series by transferring 125 μL from tube 2 to tube
3 and vortex. Repeat this step for all tubes 3-10 (see Fig. 3).
F I G U R E 3 .DOMOIC ACID STANDARD DILUTION SEQUENCE
I . PREPARATION OF SHELLFISH SAMPLES
a) Extraction of DA from shellfish samples
Shellfish flesh should be prepared as a finely blended homogenate. Preferably
analyse fresh, but it may be stored frozen at -20°C for up to 14 days before use.
1. Prepare shellfish homogenate in a high speed blender, from no less than 50 g
shellfish flesh.
2. Accuray weigh 4 g into a 50 mL centrifuge tube.
3. Add 16 mL of Extraction solution (50% methanol).
4. Mix well by vigorous shaking on vortex for 1 min.
5. Centrifuge at 3000xg for 10 minutes at room temperature.
6. Retain the supernatant for further dilution prior to analysis. The extracts can
be stored at -20˚C for up to 14 days, although with a possible reduction in DA content.
b) Dilution of shellfish sample extracts
7. Prepare dilutions of the shellfish extract in Standard/Sample buffer (10%
methanol in PBS-T) as follows:
1:20 dilution: 50 μL shellfish extract + 950 μL buffer
1:200 dilution: 50 μL of the 1:20 dilution + 450 μL buffer
1:2000 dilution: 50 μL of the 1:200 dilution + 450 μL buffer
1:20 000 dilution: 50 μL of the 1:2000 dilution + 450 μL buffer
Cap and vortex each dilution before proceeding to the next dilution step.
8. Analyze the sample dilutions according to the DA concentration range of interest
(see Table 1), to give absorbance values within the calibration curve working
range. It is recommended to analyze shellfish extracts diluted at 1:20 000
dilutions to comply with EC directive 2002/226/EC, for the quantification of DA
up to the maximum permitted level at 20 mg/kg.
T A B L E 1 : SHELLFISH EXTRACT DILUTION FOR QUANTIFICATION OF DA

J. PREPARATION OF DA SAMPLES FROM OTHER MATRICES
a) Preparation of algal samples
The analysis of algal samples will depend on the amount of algae (cells/mL) and the
amount of DA present in the algae and in the seawater or culture medium.
1. Algal samples are typically collected by gently vacuum filtering seawater
samples (10 – 100 mL) through a glass-fibre filter (GF/C, 47 mm) until the water
meniscus disappears. Do not suck hard or to dryness. To determine the amount
of DA in the seawater, also collect the flow-through filtrate and analyze this
fraction as described in “Preparation of seawater samples”.
2. Carefully lift the edge of the filter with tweezers, roll into a cylinder and insert
in a 15 mL Falcon tube. Cap and snap-freeze in dry ice as soon as possible after
filtering. The tubes can be stored at -20˚C for up to 2 weeks prior to analysis.
3. Add 10 mL of 20% methanol (1 + 4 v/v methanol plus distilled water) to the
filter in the tube.
4. Cap and vortex for 1 minute. Centrifuge at 3000xg for 10 minutes at room
temperature.
5. Dilute the algal extracts in Standard/Sample buffer before analysis. The
number of cells collected and amount of DA in the samples may vary
considerably. It is recommended that the sample dilutions tested do not
contain more than the equivalent of 100 cells/mL.
b) Preparation of seawater samples
1. Centrifuge the seawater for 10 minutes at 3000xg to remove debris from the
sample solution. Alternatively, filter the seawater through a 0.45 μm filter.
2. Dilute the cleared seawater samples in Standard/Sample buffer. For seawater
samples a minimum dilution of 1:25 is recommended.
K . ASSAY PROCEDURE
a) Incubation of standards and samples with antibody
Equilibrate pre-coated plate strips and all reagents to room temperature before use
(1 hour max). See Figure 4 for a recommended plate layout for either using 4 strips
in 2 rounds of analysis (4A), or all 8 strips at once (4B).
1. Open the packet(s) with pre-coated plate strips gently and place the strips in the
strip frame. Label each strip e.g. A, B, C and D etc.
2. Add 300 μL Washing buffer to each well. Pre-soak the wells for 5-10 minutes.
3. Remove the Washing buffer by inverting the strips over a sink and tap against
a pile of paper towels to remove all the remaining liquid.
4. Add 50 μL Standard/Sample buffer (10% methanol in PBS-T) to each of the
duplicate Amax and Blank wells.
5. Add 50 μL Antibody-HRP buffer (1% ovalbumin) to the Blank wells.
6. Add 50 μL of each DA standard dilution to each of two wells.
7. Add 50 μL of each sample dilution to each of two wells.
8. Shake vial E briefly, and tap the vial gently on a hard surface to ensure that all
the content is in the bottom of the vial. Transfer 0.5 mL (for 4 strip assay) or 1.0
mL (for 8 strip assay) from vial E (concentrated Anti-DA-HRP) to a Falcon type
tube containing 2.5 mL (for 4 strip assay) or 5.0 mL (for 8 strip assay) Antibody-
HRP buffer (1% ovalbumin). Vortex briefly.
9. Add 50 μL of the diluted Anti-DA-HRP conjugate to all wells except the Blank
wells.
10. Seal the strips with the plate sealer (B) and incubate at room temperature (20-
25°C) for 1 hour. Protect from light (e.g. cover with aluminium foil or place in
a drawer).

FIGURE 4A : SUGGESTED PLATE LAYOUT , USING 4 STRIPS, FOR THE QUANTIFICATION OF DA IN 12 SHELLFISH S AMPLES IN 2 SEPARATE ROUNDS .
S1=SAMPLE 1, S2=SAMPLE 2 , ETC.
FIGURE 4A : SUGGESTED PLATE LAYOUT , USING 8 STRIPS, FOR THE QUANTIFICATION OF DA IN 36 SHELLFISH S AMPLES 。 S1=SAMPLE 1, S2=SAMPLE 2 , ETC.
b) Developing and reading the microplate strips
11. Carefully remove the plate sealer. Wash the wells 4 times with 300 μL Washing
buffer per well.
12. Add 100 μL of TMB peroxidase substrate (vial G) to all wells. Incubate at room
temperature (20-25°C) for 15 minutes. Protect from light.
13. Stop the reaction by adding 100 μL 0.3 M H2SO4 to all wells.
14. After 2-5 minutes, read the absorbance in a microplate spectrophotometer
using a 450 nm filter.
L . CALCULATION OF RESULTS
a) Calibration using the four-parameter logistic curve fit model
When the measured absorbance values of the standard dilutions are plotted on a
linear scale (y axis) against the DA-concentrations of the standard dilutions on a
logarithmic scale (x axis), a sigmoid (S-shaped) curve is obtained (see Fig. 5).
The non-linear 4-parameter logistic curve-fit model is extensively used for sigmoid
curves, in order to get accurate quantification of samples and a good fit at the
extremes of the curve. The following equation is given for a 4-parameter fitted
curve:
y = (a-d)/[1+(x/c)b]+d
where:
x is the concentration of DA in the standard/sample
y is the absorbance of the standard/sample
a is the y-value of the upper asymptote (Amax)
b is the relative slope of the curve at its center
c is the x-value at the midpoint of the curve (I50)
d is the y-value of the lower asymptote (Blank/Amin)

F I G U R E 5 .
NON-LINEAR CALIBRA-TION CURVE PREPARED BY 4-PARAMETER LO-GISTIC CURVE FIT .
b) Calculation formula
The following formula is used to convert ELISA results in pg/mL to shellfish
concentrations in mg/kg:
mg DA/kg = μg DA/g = 1 000 000 pg DA/mL · D · V / M
where:
pg DA/mL is the concentration of DA in the diluted extract
D is the dilution factor of the diluted extract
V is the volume of the methanolic extract (16 mL plus 4 g of homogenate giving nominal
20 mL total volume).
M is the mass of the shellfish homogenate (4 g).
c) Excel macro EMA31 calculation of DA concentration in shellfish samples
For calculation of assay results, a spreadsheet has been developed implementing
the calibration function and the conversion formula from pg/mL in the extract to
mg DA/kg shellfish.
1. Open the provided Excel Macro EMA31, enable macros and install the Solver
as described in the “Instructions” sheet of the Macro.
2. Copy the measured absorbance values (to at least 3 significant figures, e.g.
0.682) from the plate reading software result/report sheet and paste the values
in the Excel Macro EMA31 “Data Entry” sheet.
3. Enter the correct dilution factor used for the samples, in the corresponding
duplicate well windows.
4. Run the macro according to the instructions.
5. Go to the “Results” sheet. The results from the column “Shellfish sample DA
eqv. (mg/kg)” give the concentration of DA in the shellfish samples.
6. Sample concentrations should only be calculated from datapoints that are
within the valid working range of the standard curve as defined by the Excel
macro. If more than one sample dilution hit the working range of the standard
curve, we recommend that the dilution closest to the I50 value of the standard
is used.
Alternatively; another data analysis software (e.g. the software provided with
the plate reader) may be used as long as it supports the 4-parameter logistic
curve fit model.
d) Excel macro EMA31 calculation of DA concentration in Algal samples
1. Use the provided Excel macro EMA31 as described in the previous section.
2. Enter the correct dilution factor used for the algal samples, in the corresponding
duplicate well windows.
3. The results from the column “Sample extract/solution (pg/mL)” will provide the DA
concentration of the algae extracts as pg/mL.
4. If Pseudo-nitzschia cell counts are available for the filtered water sample, the
results can be converted to pg DA/cell, taking into account the volume of water
filtered and the extraction volume.
e) Excel macro calculation of DA concentration in seawater samples
1. Use the provided Excel macro EMA31 as described in the previous section.
2. Enter the correct dilution factor used for the seawater samples, in the
corresponding duplicate well windows.
3. The results from the column “Sample extract/solution (pg/mL)” will provide the DA
concentration of the seawater samples as pg/mL.
M . QUALITY ASSURANCE MEASURES FOR VALID ANALYSIS
• In order to qualify as a valid calibration curve suitable for accurate quantification
of DA in samples, the requirements listed in Table 2 must be fulfilled.
• Sample concentrations should only be calculated from datapoints that are
within the valid working range of the calibration curve as defined by the Excel macro.
• The estimated curve fit (%CV of prediction) for the calibration curve should be
<20%, as indicated in the “Results” sheet of the Excel Macro EMA31.
• The concentration difference should not be more 15% between two duplicate
wells for a given sample.
TABLE2 : QUALITY ASSURANCE REQUIREMENTS FOR VALID CALIBRATION CURVE

N . REFERENCES
• Garthwaite I, Ross KM, Miles CO, Briggs L, Towers N, Borell T, Busby P. (2001) J. AOAC Int.
84, 1643-1648.
• Garthwaite I, Ross KM, Miles CO, Hansen RP, Foster D, Wilkins, AL, Towers N. (1998) Nat.
Toxins 6, 93-104.
• Kleivdal H, Holland P, McNabb P. (2003) Biosense Laboratories AS report No. 2003:6.
• Kleivdal H. (2004) Biosense Laboratories AS report No. 2004:1.2.
• Scholin CA, Gulland F, Douchette GJ et al. (2000) Nature 403, 80-84.
• Wright JLC, Boyd RK, DeFreitas ASW, Falk M, Foxall RA, Jamieson WD, Laycock MV,
McCulloch AW, McInnes AG, Odense P, Pathak VP, Quilliam MA, Ragan MA, Sim PG,
Thibault P, Walter JA, Richard DJA, Dwar D. (1989) Can. J. Chem. 67, 481-490.
O . QUICKGUIDE
1. Prepare dilutions of standard and samples.
2. Pre-soak the wells for 5-10 minutes with 300 μl Washing buffer. Empty before
use.
3. Add 50 μL Standard/Sample buffer to the Amax and Blank wells.
4. Add 50 μl Antibody-HRP buffer to the Blank wells.
5. Transfer 50 μL of diluted standards and samples (in duplicate) to the plate.
6. Dilute the concentrated antibody-HRP conjugate and add 50 μL to all wells
except the Blank wells.
7. Seal the plate and incubate at room temperature for 1 hour (keep dark).
8. Wash the wells.
9. Add 100 μL TMB peroxidase substrate to all wells.
10. Incubate at room temperature for 15 minutes (keep dark).
11. Add 100 μL of 0.3 M H2SO4 to all wells to stop the reaction.
12. Read the absorbance at 450 nm.
13. Calculate the concentrations using the Excel Macro EMA31.


 BS31300401
BS31300401 96
96